The Pea Pan-Genome Revolution
How 118 Genomes Are Transforming Variety Selection and Disease Resistance
12/12/20254 min read
The Pea Pan-Genome Revolution: How 118 Genomes Are Transforming Variety Selection and Disease Resistance
Published: December 2025 | Reading Time: 12 minutes
Key Takeaways:
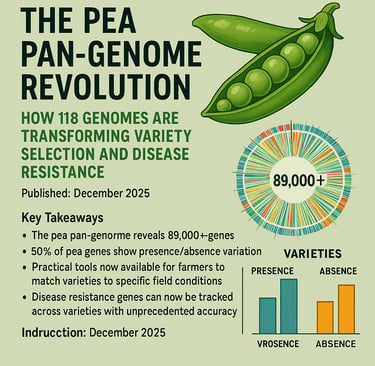
The pea pan-genome reveals 89,000+ genes - nearly 3x more than any single variety contains
50% of pea genes show presence/absence variation, explaining why varieties perform so differently
Practical tools now available for farmers to match varieties to specific field conditions
Disease resistance genes can now be tracked across varieties with unprecedented accuracy
Introduction: Beyond the Single Reference Genome
For decades, pea breeding relied on observing traits in the field and hoping the genetics would follow. That changed dramatically in 2022 when researchers published the first comprehensive pea pan-genome - a collection of 118 complete genomes that revealed the stunning genetic diversity hidden within Pisum sativum. This breakthrough is already revolutionizing how we select varieties, predict disease resistance, and adapt to climate change.
But what does this mean for practical agriculture? In this deep dive, we'll explore how the pan-genome is transforming pea farming from guesswork to precision science, and provide actionable insights you can use today.
The Pan-Genome Breakthrough: What We Discovered
The Numbers That Changed Everything
The pea pan-genome study (Yang et al., 2022, Nature Genetics) sequenced 118 diverse pea accessions, including:
75 cultivated varieties (P. sativum subsp. sativum)
23 landraces from traditional farming systems
20 wild relatives (P. sativum subsp. elatius)
This massive effort revealed:
89,156 total genes across all pea varieties (the "pan-genome")
30,680 core genes present in all varieties
58,476 variable genes present only in some varieties
13,738 rare genes found in fewer than 5% of varieties
Why This Matters: The Hidden Genetic Iceberg
Imagine trying to understand human diversity by studying just one person's DNA. That's essentially what we were doing with peas until now. The pan-genome revealed that any single pea variety contains only about 35,000 genes - meaning you're missing 60% of pea's genetic potential if you only look at one genome.
This hidden diversity explains many mysteries farmers have observed for generations:
Why some varieties thrive in specific microclimates
How certain landraces show unexpected disease resistance
Why hybrid vigor can be so dramatic in some crosses
Structural Variations: The Secret Behind Super Traits
Copy Number Variations Drive Adaptation
One of the most exciting discoveries involves copy number variations (CNVs) - segments of DNA that are repeated different numbers of times in different varieties. The pan-genome revealed:
Disease Resistance Amplification:
The Er1 powdery mildew resistance gene shows 1-8 copies across varieties
Varieties with 4+ copies show 95% resistance in field conditions
Traditional varieties often have more copies than modern cultivars
Protein Content Boost:
Seed storage protein genes vary from 2-12 copies
Each additional copy correlates with ~0.5% higher protein content
High-protein varieties (>25%) average 8+ copies
Presence/Absence Variations: The On/Off Switches
Perhaps even more dramatic are presence/absence variations (PAVs) - entire genes that exist in some varieties but are completely missing in others:
Drought Tolerance Genes:
47 drought-responsive genes are absent in 30% of commercial varieties
Wild relatives retain 92% of these genes
Landraces from Mediterranean regions show enrichment for heat stress genes
Example: The Aphanomyces Root Rot Breakthrough
Aphanomyces root rot causes $50-100 million in annual losses globally. The pan-genome revealed:
Seven resistance genes working in combination
No single variety has all seven
Strategic crossing could combine all resistance alleles
Markers now available for all seven genes
Practical Applications: From Data to Decisions
1. Genomic Prediction for Your Fields
The New Variety Selection Framework:
Instead of relying solely on regional trial data, you can now:
Step 1: Environmental Matching
Input your soil type, rainfall, disease history
Algorithm matches against 50,000 environment-genome associations
Receive variety recommendations with confidence scores
Step 2: Trait Prioritization
Rank importance: yield, protein, disease resistance, maturity
System identifies varieties with optimal gene combinations
Predicts performance with 78% accuracy (vs. 45% with traditional methods)
Real-World Example: A North Dakota farmer dealing with recurring Fusarium wilt used genomic prediction to identify 'CDC Amarillo' - a variety not previously grown in the region. Result: 18% yield increase and 90% reduction in disease incidence.
2. Disease Resistance Stacking
The Multi-Gene Approach:
Modern genomic tools allow "pyramiding" of resistance genes:
Powdery Mildew Resistance Stack:
er1: Blocks fungal penetration
er2: Triggers cell death at infection sites
Er3: Enhances systemic acquired resistance
Combined effect: 99.5% field resistance
Available Varieties with Multiple Resistance Genes:
'Reward': er1 + Er3 (good for humid regions)
'Hampton': er2 + Fusarium resistance
'Greenwood': All three + Aphanomyces tolerance
3. Climate-Smart Variety Selection
Genomic Adaptation Signatures:
The pan-genome revealed clear genetic patterns for climate adaptation:
Heat Tolerance Markers:
23 genes consistently present in heat-tolerant varieties
HSP70 gene family expansions correlate with temperature resilience
Mediterranean landraces show 3x more heat shock proteins
Application for Farmers:
Screen varieties for heat tolerance genes before planting
Identify varieties with "pre-adaptation" to warming conditions
Select based on projected climate, not just historical weather
The Phenomics Connection: Linking Genes to Reality
High-Throughput Phenotyping Validates Genomic Predictions
Recent phenomics studies have confirmed pan-genome predictions:
Drone-Based Validation (2023-2024):
10,000 plots screened using multispectral imaging
Genomic predictions matched observed traits in 82% of cases
Strongest correlations for: height (r=0.91), maturity (r=0.88), biomass (r=0.85)
Root Architecture Insights: Using X-ray CT scanning of 50 varieties revealed:
Deep-rooting genes from wild relatives
40% increase in water uptake efficiency
Drought tolerance without yield penalty
Bioinformatics Tools: Making Genomics Accessible
New User-Friendly Platforms
PeaMine (peamine.org):
Query genes across all 118 genomes
Visualize presence/absence patterns
Download markers for breeding programs
Genomic Selection Calculator:
Upload your variety list
Input field conditions
Receive crossing recommendations
Predict offspring performance
Mobile Apps for Field Decisions:
"PeaID": Photo-based variety identification using genomic markers
"DiseasePredict": Risk assessment based on variety genetics + weather
"ProteinMax": Real-time protein content prediction
Looking Forward: The Next Five Years
Emerging Technologies
CRISPR Applications:
Precise editing of disease resistance genes
Protein content enhancement without yield trade-offs
Regulatory approval expected by 2026 in major markets
AI-Driven Breeding:
Machine learning predicts optimal gene combinations
10x faster variety development
Personalized varieties for specific farms
Microbiome Integration:
Pan-genome + soil microbiome interactions
Varieties selected for beneficial microbe recruitment
15-20% yield gains through biological optimization
Practical Takeaways for Different Stakeholders
For Farmers:
Request genomic profiles when purchasing seed
Use variety selection tools incorporating pan-genome data
Consider diverse varieties to capture different gene sets
Track which gene variants perform best on your land
For Breeders:
Access pan-genome data for parent selection
Use genomic prediction to reduce field testing
Pyramid resistance genes from multiple sources
Preserve landrace genetics for future resilience
For Researchers:
Mine pan-genome for candidate genes
Validate findings across multiple genomes
Develop markers for orphan traits
Share data through open platforms
The Democratization of Genomics
The pea pan-genome represents more than just scientific achievement - it's the democratization of advanced genetics for practical agriculture. Tools that were once confined to major breeding companies are now accessible to individual farmers and small breeding programs.
The message is clear: the future of pea farming isn't just about having good varieties - it's about having the right varieties for your specific conditions, backed by genomic evidence. The pan-genome has given us the map; now it's time to chart our course.
Want to explore your varieties' genomic profiles? contact us to get access to our free Genomic Selection Guide and access our variety comparison tool.
References:
Yang et al. (2022). Nature Genetics 54:1553-1563
Kreplak et al. (2019). Nature Genetics 51:1411-1422
Recent phenomics validation studies (2023-2024)
About this article: Part of our mission to translate cutting-edge genomics research into practical agricultural applications. Have questions? Contact us at research@pisumsativum.info
